Curate datasets¶
Curating a dataset with LaminDB means three things:
Validate that the dataset matches a desired schema
If validation fails, standardize the dataset (e.g., by fixing typos, mapping synonyms) or update registries
Annotate the dataset by linking it against metadata entities so that it becomes queryable
In this guide we’ll curate common data structures. Here is a guide for the underlying low-level API.
Note: If you know either pydantic or pandera, here is an FAQ that compares LaminDB with both of these tools.
# pip install 'lamindb[bionty]'
!lamin init --storage ./test-curate --modules bionty
import lamindb as ln
ln.track("MCeA3reqZG2e")
DataFrame¶
Allow a flexible schema¶
We’ll be working with the mini immuno dataset:
df = ln.core.datasets.mini_immuno.get_dataset1()
df
This is how we curate it in a script.
import lamindb as ln
ln.core.datasets.mini_immuno.define_features_labels()
schema = ln.examples.schemas.valid_features()
df = ln.core.datasets.small_dataset1(otype="DataFrame")
artifact = ln.Artifact.from_df(
df, key="examples/dataset1.parquet", schema=schema
).save()
artifact.describe()
Let’s run the script.
!python scripts/curate_dataframe_flexible.py
The script defined the following features & labels through define_features_labels():
import lamindb as ln
import bionty as bt
# define valid labels
perturbation_type = ln.ULabel(name="Perturbation", is_type=True).save()
ln.ULabel(name="DMSO", type=perturbation_type).save()
ln.ULabel(name="IFNG", type=perturbation_type).save()
bt.CellType.from_source(name="B cell").save()
bt.CellType.from_source(name="T cell").save()
# define valid features
ln.Feature(name="perturbation", dtype=perturbation_type).save()
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save()
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save()
ln.Feature(name="assay_oid", dtype=bt.ExperimentalFactor.ontology_id).save()
ln.Feature(name="concentration", dtype=str).save()
ln.Feature(name="treatment_time_h", dtype="num", coerce_dtype=True).save()
ln.Feature(name="donor", dtype=str, nullable=True).save()
ln.Feature(name="donor_ethnicity", dtype=list[bt.Ethnicity]).save()
And the following schema through valid_features():
import lamindb as ln
schema = ln.Schema(name="valid_features", itype=ln.Feature).save()
Require a minimal set of columns¶
If we’d like to curate the dataframe with a minimal set of required columns, we can use the following schema.
import lamindb as ln
schema = ln.Schema(
name="Mini immuno schema",
features=[
ln.Feature.get(name="perturbation"),
ln.Feature.get(name="cell_type_by_model"),
ln.Feature.get(name="assay_oid"),
ln.Feature.get(name="donor"),
ln.Feature.get(name="concentration"),
ln.Feature.get(name="treatment_time_h"),
],
flexible=True, # _additional_ columns in a dataframe are validated & annotated
).save()
If the dataframe lacks one of the required columns, we’ll get a validation error.
import lamindb as ln
schema = ln.core.datasets.mini_immuno.define_mini_immuno_schema_flexible()
df = ln.core.datasets.small_dataset1(otype="DataFrame")
df.pop("donor") # remove donor column to trigger validation error
try:
artifact = ln.Artifact.from_df(
df, key="examples/dataset1.parquet", schema=schema
).save()
except ln.errors.ValidationError as error:
print(error)
Let’s run the script.
!python scripts/curate_dataframe_minimal_errors.py
Resolve synonyms and typos¶
Let’s now look at the same dataset but assume there are synonyms and typos.
df = ln.core.datasets.mini_immuno.get_dataset1(
with_cell_type_synonym=True, with_cell_type_typo=True
)
df
Let’s reuse the schema that defines a minimal set of columns we expect in the dataframe.
schema = ln.core.datasets.mini_immuno.define_mini_immuno_schema_flexible()
schema.describe()
Create a curator object using the dataset & the schema.
curator = ln.curators.DataFrameCurator(df, schema)
The validate() method validates that your dataset adheres to the criteria defined by the schema. It identifies which values are already validated (exist in the registries) and which are potentially problematic (do not yet exist in our registries).
try:
curator.validate()
except ln.errors.ValidationError as error:
print(error)
# check the non-validated terms
curator.cat.non_validated
For cell_type, we saw that “cerebral pyramidal neuron”, “astrocytic glia” are not validated.
First, let’s standardize synonym “astrocytic glia” as suggested
curator.cat.standardize("cell_type_by_expert")
# now we have only one non-validated cell type left
curator.cat.non_validated
For “CD8-pos alpha-beta T cell”, let’s understand which cell type in the public ontology might be the actual match.
# to check the correct spelling of categories, pass `public=True` to get a lookup object from public ontologies
# use `lookup = curator.cat.lookup()` to get a lookup object of existing records in your instance
lookup = curator.cat.lookup(public=True)
lookup
# here is an example for the "cell_type" column
cell_types = lookup["cell_type_by_expert"]
cell_types.cd8_positive_alpha_beta_t_cell
# fix the cell type name
df["cell_type_by_expert"] = df["cell_type_by_expert"].cat.rename_categories(
{"CD8-pos alpha-beta T cell": cell_types.cd8_positive_alpha_beta_t_cell.name}
)
For perturbation, we want to add the new values: “DMSO”, “IFNG”
# this adds perturbations that were _not_ validated
curator.cat.add_new_from("perturbation")
# validate again
curator.validate()
Save a curated artifact.
artifact = curator.save_artifact(key="examples/my_curated_dataset.parquet")
artifact.describe()
AnnData¶
AnnData like all other data structures that follow is a composite structure that stores different arrays in different slots.
Allow a flexible schema¶
We can also allow a flexible schema for an AnnData and only require that it’s indexed with Ensembl gene IDs.
import lamindb as ln
ln.core.datasets.mini_immuno.define_features_labels()
adata = ln.core.datasets.mini_immuno.get_dataset1(otype="AnnData")
schema = ln.examples.schemas.anndata_ensembl_gene_ids_and_valid_features_in_obs()
artifact = ln.Artifact.from_anndata(
adata, key="examples/mini_immuno.h5ad", schema=schema
).save()
artifact.describe()
Let’s run the script.
!python scripts/curate_anndata_flexible.py
Under-the-hood, this used the following schema:
import lamindb as ln
import bionty as bt
obs_schema = ln.examples.schemas.valid_features()
varT_schema = ln.Schema(
name="valid_ensembl_gene_ids", itype=bt.Gene.ensembl_gene_id
).save()
schema = ln.Schema(
name="anndata_ensembl_gene_ids_and_valid_features_in_obs",
otype="AnnData",
slots={"obs": obs_schema, "var.T": varT_schema},
).save()
This schema tranposes the var DataFrame during curation, so that one validates and annotates the var.T schema, i.e., [ENSG00000153563, ENSG00000010610, ENSG00000170458].
If one doesn’t transpose, one would annotate with the schema of var, i.e., [gene_symbol, gene_type].
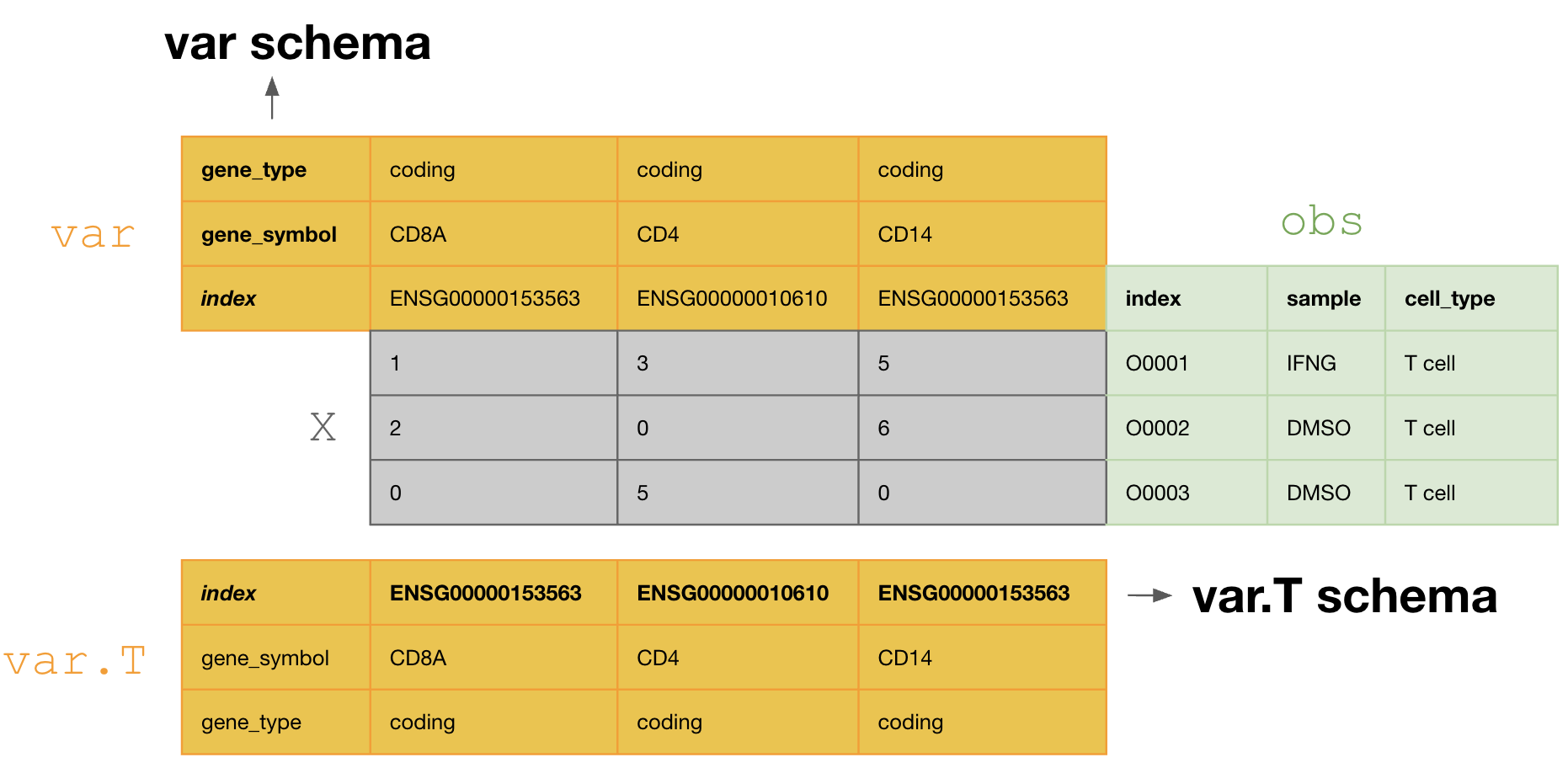
Resolve typos¶
import lamindb as ln
adata = ln.core.datasets.mini_immuno.get_dataset1(
with_gene_typo=True, with_cell_type_typo=True, otype="AnnData"
)
adata
Check the slots of a schema:
schema.slots
curator = ln.curators.AnnDataCurator(adata, schema)
try:
curator.validate()
except ln.errors.ValidationError as error:
print(error)
As above, we leverage a lookup object with valid cell types to find the correct name.
valid_cell_types = curator.slots["obs"].cat.lookup()["cell_type_by_expert"]
adata.obs["cell_type_by_expert"] = adata.obs[
"cell_type_by_expert"
].cat.rename_categories(
{"CD8-pos alpha-beta T cell": valid_cell_types.cd8_positive_alpha_beta_t_cell.name}
)
The validated AnnData can be subsequently saved as an Artifact:
adata.obs.columns
Index(['perturbation', 'sample_note', 'cell_type_by_expert',
'cell_type_by_model', 'assay_oid', 'concentration', 'treatment_time_h',
'donor'],
dtype='object')
curator.slots["var.T"].cat.add_new_from("columns")
! using default organism = human
! 1 term not validated in feature 'columns' in slot 'var.T': 'GeneTypo'
→ fix typos, remove non-existent values, or save terms via: curator.slots['var.T'].cat.add_new_from('columns')
curator.validate()
! 1 term not validated in feature 'columns' in slot 'obs': 'sample_note'
→ fix typos, remove non-existent values, or save terms via: curator.slots['obs'].cat.add_new_from('columns')
artifact = curator.save_artifact(key="examples/my_curated_anndata.h5ad")
Access the schema for each slot:
artifact.features.slots
The saved artifact has been annotated with validated features and labels:
artifact.describe()
MuData¶
import lamindb as ln
import bionty as bt
# define the global obs schema
obs_schema = ln.Schema(
name="mudata_papalexi21_subset_obs_schema",
features=[
ln.Feature(name="perturbation", dtype="cat[ULabel[Perturbation]]").save(),
ln.Feature(name="replicate", dtype="cat[ULabel[Replicate]]").save(),
],
).save()
# define the ['rna'].obs schema
obs_schema_rna = ln.Schema(
name="mudata_papalexi21_subset_rna_obs_schema",
features=[
ln.Feature(name="nCount_RNA", dtype=int).save(),
ln.Feature(name="nFeature_RNA", dtype=int).save(),
ln.Feature(name="percent.mito", dtype=float).save(),
],
).save()
# define the ['hto'].obs schema
obs_schema_hto = ln.Schema(
name="mudata_papalexi21_subset_hto_obs_schema",
features=[
ln.Feature(name="nCount_HTO", dtype=int).save(),
ln.Feature(name="nFeature_HTO", dtype=int).save(),
ln.Feature(name="technique", dtype=bt.ExperimentalFactor).save(),
],
).save()
# define ['rna'].var schema
var_schema_rna = ln.Schema(
name="mudata_papalexi21_subset_rna_var_schema",
itype=bt.Gene.symbol,
dtype=float,
).save()
# define composite schema
mudata_schema = ln.Schema(
name="mudata_papalexi21_subset_mudata_schema",
otype="MuData",
slots={
"obs": obs_schema,
"rna:obs": obs_schema_rna,
"hto:obs": obs_schema_hto,
"rna:var": var_schema_rna,
},
).save()
# curate a MuData
mdata = ln.core.datasets.mudata_papalexi21_subset()
bt.settings.organism = "human" # set the organism to map gene symbols
curator = ln.curators.MuDataCurator(mdata, mudata_schema)
artifact = curator.save_artifact(key="examples/mudata_papalexi21_subset.h5mu")
assert artifact.schema == mudata_schema
!python scripts/curate_mudata.py
SpatialData¶
import lamindb as ln
import bionty as bt
attrs_schema = ln.Schema(
features=[
ln.Feature(name="bio", dtype=dict).save(),
ln.Feature(name="tech", dtype=dict).save(),
],
).save()
sample_schema = ln.Schema(
features=[
ln.Feature(name="disease", dtype=bt.Disease, coerce_dtype=True).save(),
ln.Feature(
name="developmental_stage",
dtype=bt.DevelopmentalStage,
coerce_dtype=True,
).save(),
],
).save()
tech_schema = ln.Schema(
features=[
ln.Feature(name="assay", dtype=bt.ExperimentalFactor, coerce_dtype=True).save(),
],
).save()
obs_schema = ln.Schema(
features=[
ln.Feature(name="sample_region", dtype="str").save(),
],
).save()
# Schema enforces only registered Ensembl Gene IDs are valid (maximal_set=True)
varT_schema = ln.Schema(itype=bt.Gene.ensembl_gene_id, maximal_set=True).save()
sdata_schema = ln.Schema(
name="spatialdata_blobs_schema",
otype="SpatialData",
slots={
"attrs:bio": sample_schema,
"attrs:tech": tech_schema,
"attrs": attrs_schema,
"tables:table:obs": obs_schema,
"tables:table:var.T": varT_schema,
},
).save()
!python scripts/define_schema_spatialdata.py
import lamindb as ln
spatialdata = ln.core.datasets.spatialdata_blobs()
sdata_schema = ln.Schema.get(name="spatialdata_blobs_schema")
curator = ln.curators.SpatialDataCurator(spatialdata, sdata_schema)
try:
curator.validate()
except ln.errors.ValidationError:
pass
spatialdata.tables["table"].var.drop(index="ENSG00000999999", inplace=True)
# validate again (must pass now) and save artifact
artifact = ln.Artifact.from_spatialdata(
spatialdata, key="examples/spatialdata1.zarr", schema=sdata_schema
).save()
artifact.describe()
!python scripts/curate_spatialdata.py
TiledbsomaExperiment¶
import lamindb as ln
import bionty as bt
import tiledbsoma as soma
import tiledbsoma.io
adata = ln.core.datasets.mini_immuno.get_dataset1(otype="AnnData")
tiledbsoma.io.from_anndata("small_dataset.tiledbsoma", adata, measurement_name="RNA")
obs_schema = ln.Schema(
name="soma_obs_schema",
features=[
ln.Feature(name="cell_type_by_expert", dtype=bt.CellType).save(),
ln.Feature(name="cell_type_by_model", dtype=bt.CellType).save(),
],
).save()
var_schema = ln.Schema(
name="soma_var_schema",
features=[
ln.Feature(name="var_id", dtype=bt.Gene.ensembl_gene_id).save(),
],
coerce_dtype=True,
).save()
soma_schema = ln.Schema(
name="soma_experiment_schema",
otype="tiledbsoma",
slots={
"obs": obs_schema,
"ms:RNA.T": var_schema,
},
).save()
with soma.Experiment.open("small_dataset.tiledbsoma") as experiment:
curator = ln.curators.TiledbsomaExperimentCurator(experiment, soma_schema)
curator.validate()
artifact = curator.save_artifact(
key="examples/soma_experiment.tiledbsoma",
description="SOMA experiment with schema validation",
)
assert artifact.schema == soma_schema
artifact.describe()
!python scripts/curate_soma_experiment.py
Other data structures¶
If you have other data structures, read: How do I validate & annotate arbitrary data structures?.